scRNA-seq of human vitiligo reveals complex networks of subclinical immune activation and a role for CCR5 in Treg function
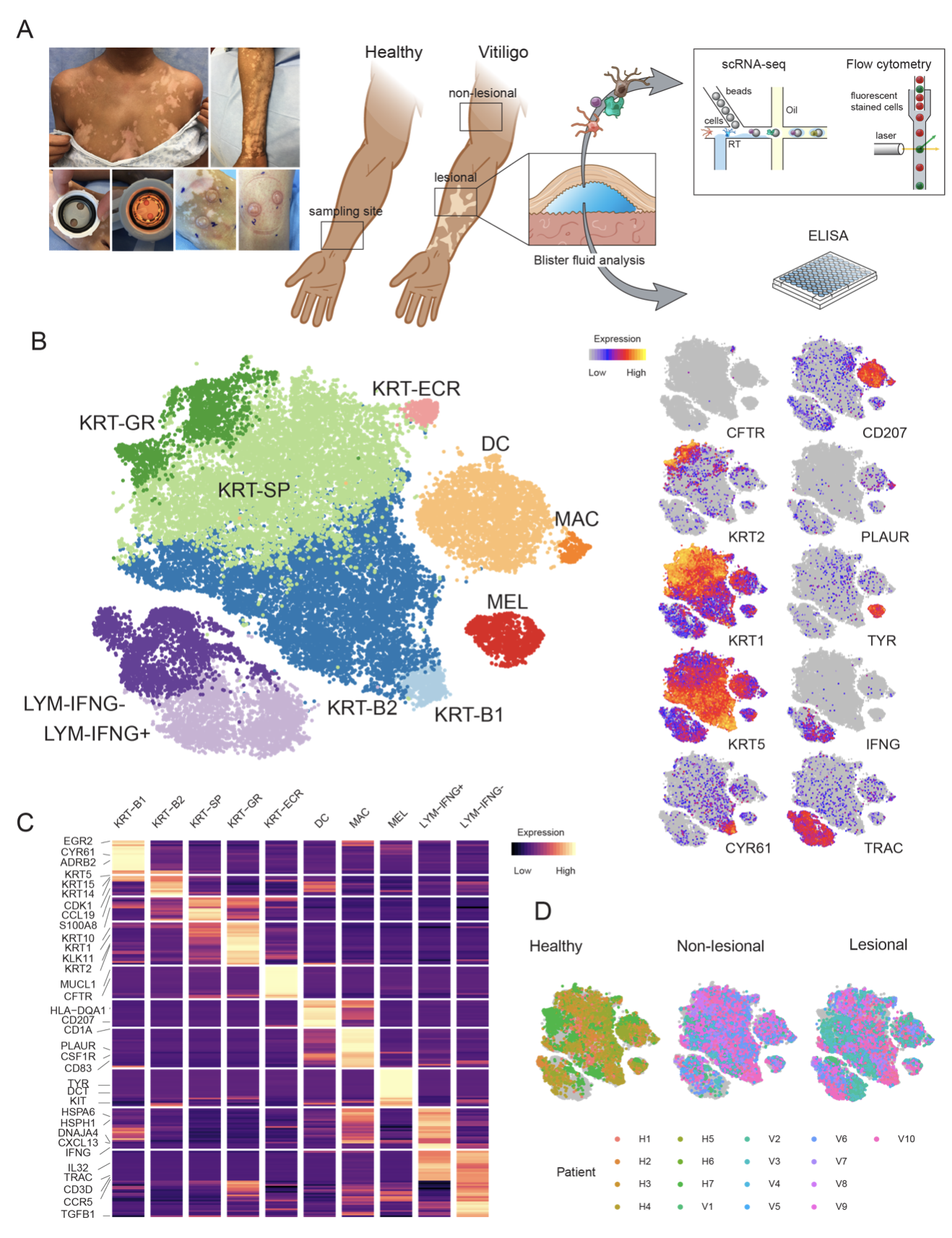
This site provides access to the data we used in our recent publication (Gellatly et. al 2021) where we sought to identify cellular communications that show disruption in lesions or in non-lesional skins of individuals suffering from Vitiligo, an autoimmune skin disease characterized by the targeted destruction of melanocytes by T cells.
The data provided was obtained using our in-house inDrop system to generate single cell RNA-Seq profiles on affected and unaffected skin from vitiligo patients, as well as healthy controls. The data was analyzed using our end-to-end scRNA data analysis package called SignallingSingleCell.
To provide access to the processed data we are hosting an interactive browser based on Cellxgene Browser.
All raw sequencing data was processed by the scRNA-Seq inDrop pipeline developed within the interactive pipeline manager DolphinNext (Yukselen et. al 2020), and currently being hosted in DolphinSuite high throughput data analysis platform.
Raw Data:
The raw .fastq files have been deposited through dbGAP. These files are protected, and you will need an account and to request access to download the files. As of 10/5/21 the data files are processing on the dbGAP server.
https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002455.v1.p1
UMI Tables:
For convenience we are providing 3 processed data tables here. One is a raw sparse matrix, that is the output of our processing pipeline with no further manipulation. We are also providing a fully processed UMI table that has been filtered, dimension reduced, clustered, and normalized. The processed UMI table is provided in the ExpressionSet class format. We are also providing a separate processed UMI table for the T Cells.
Processed UMI Table of T-cells